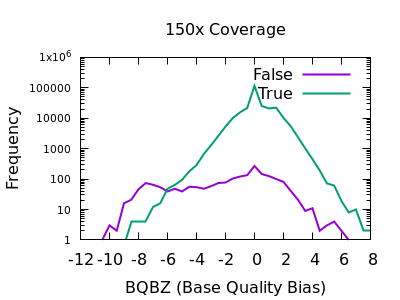
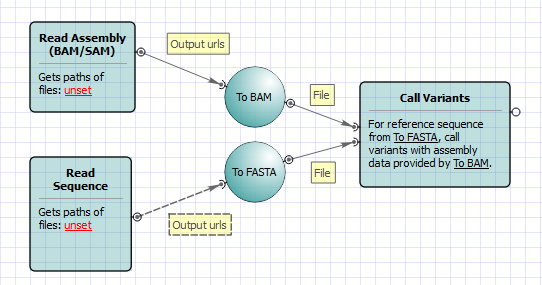
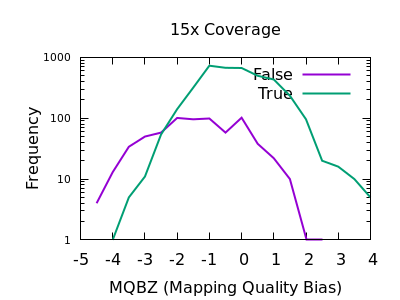
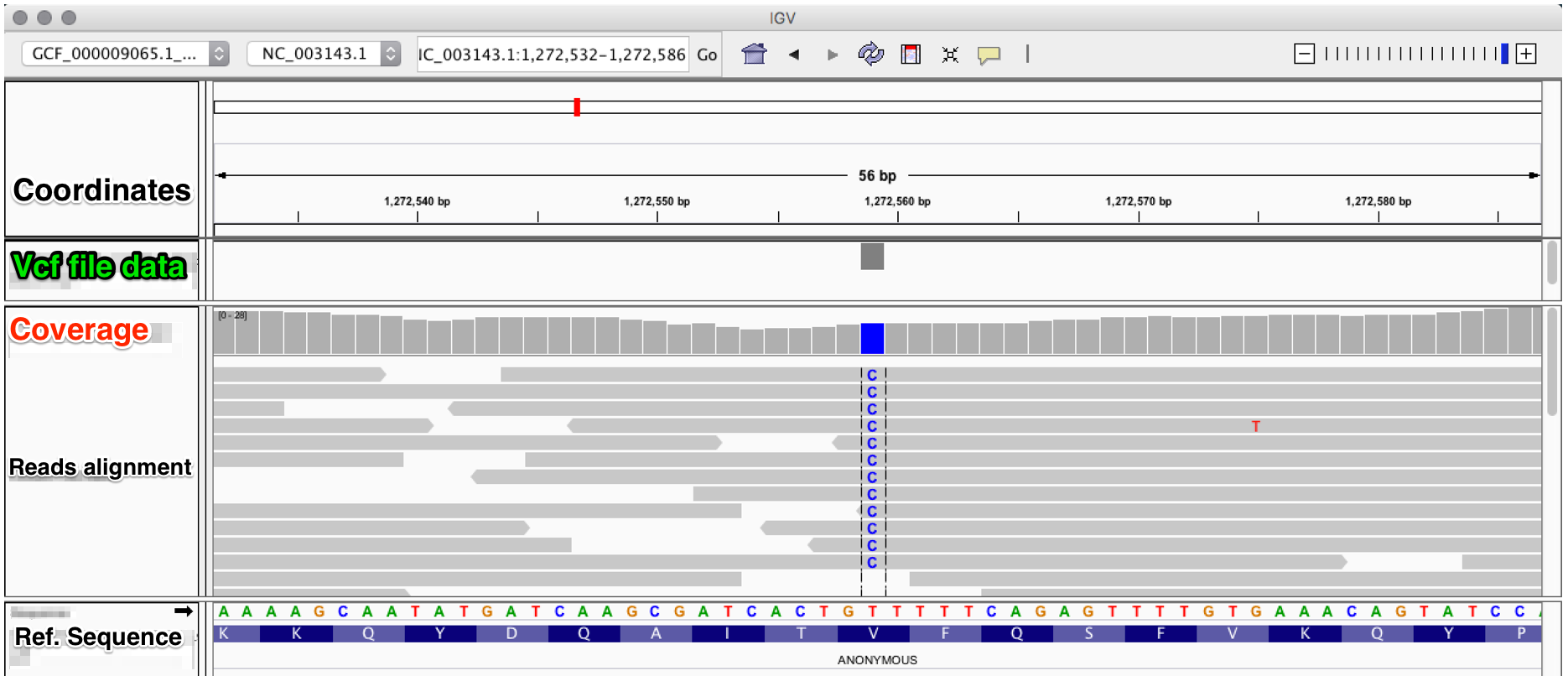
bcftools filter -e 'GT="het"' recognize GT 2/2 as heterozygous · Issue #1268 · samtools/bcftools · GitHub

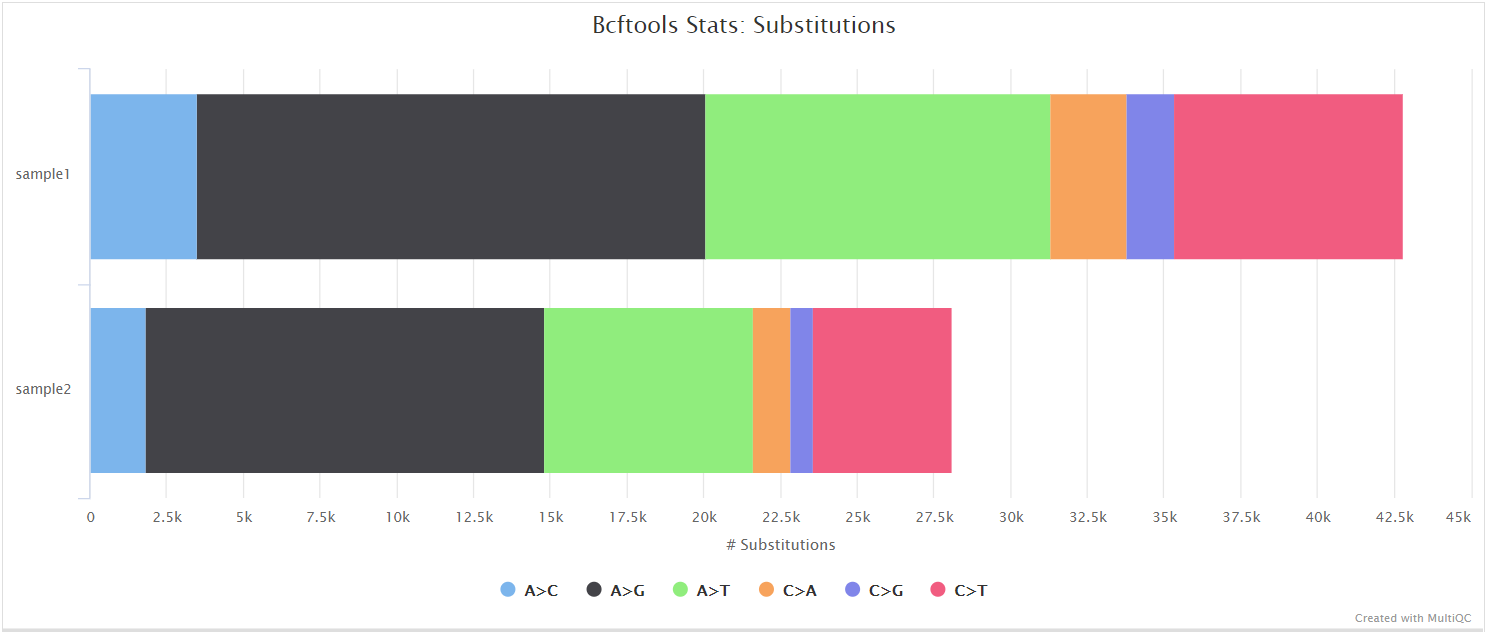
bcftools mpileup - Difference between IDV and FORMAT/AD* fields · Issue #912 · samtools/bcftools · GitHub
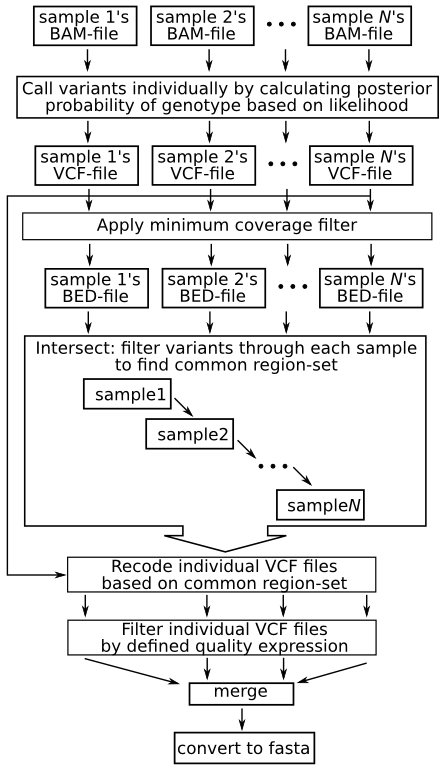
a) Filtering different variant callers VCF output for SARS-CoV-2 data.... | Download Scientific Diagram

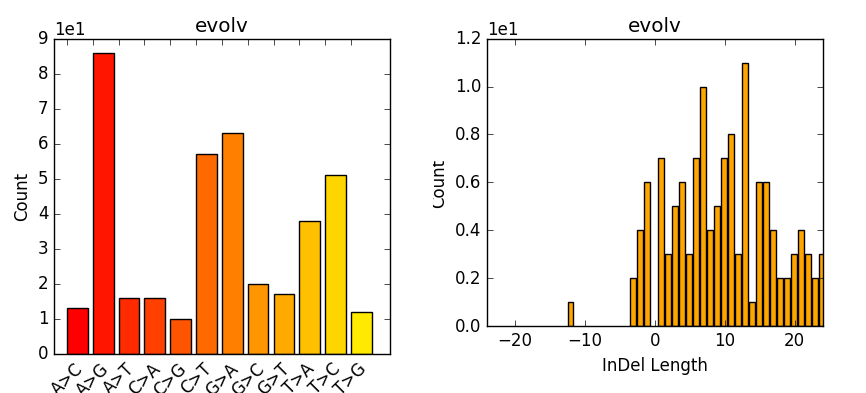
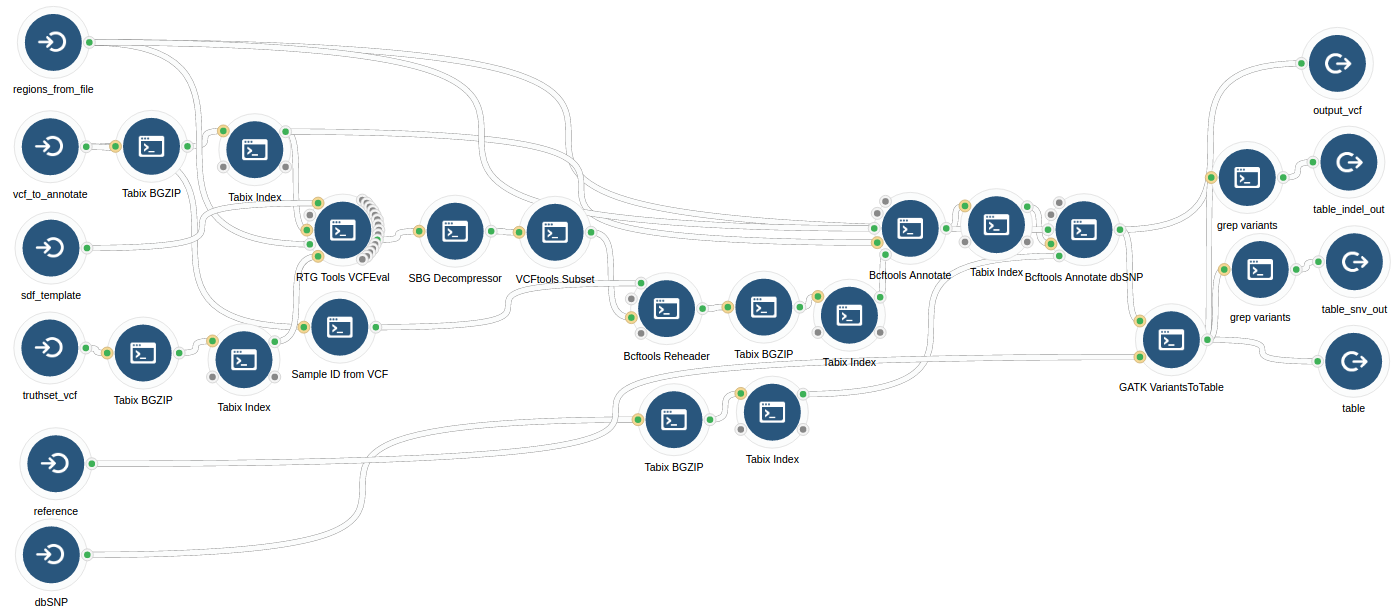
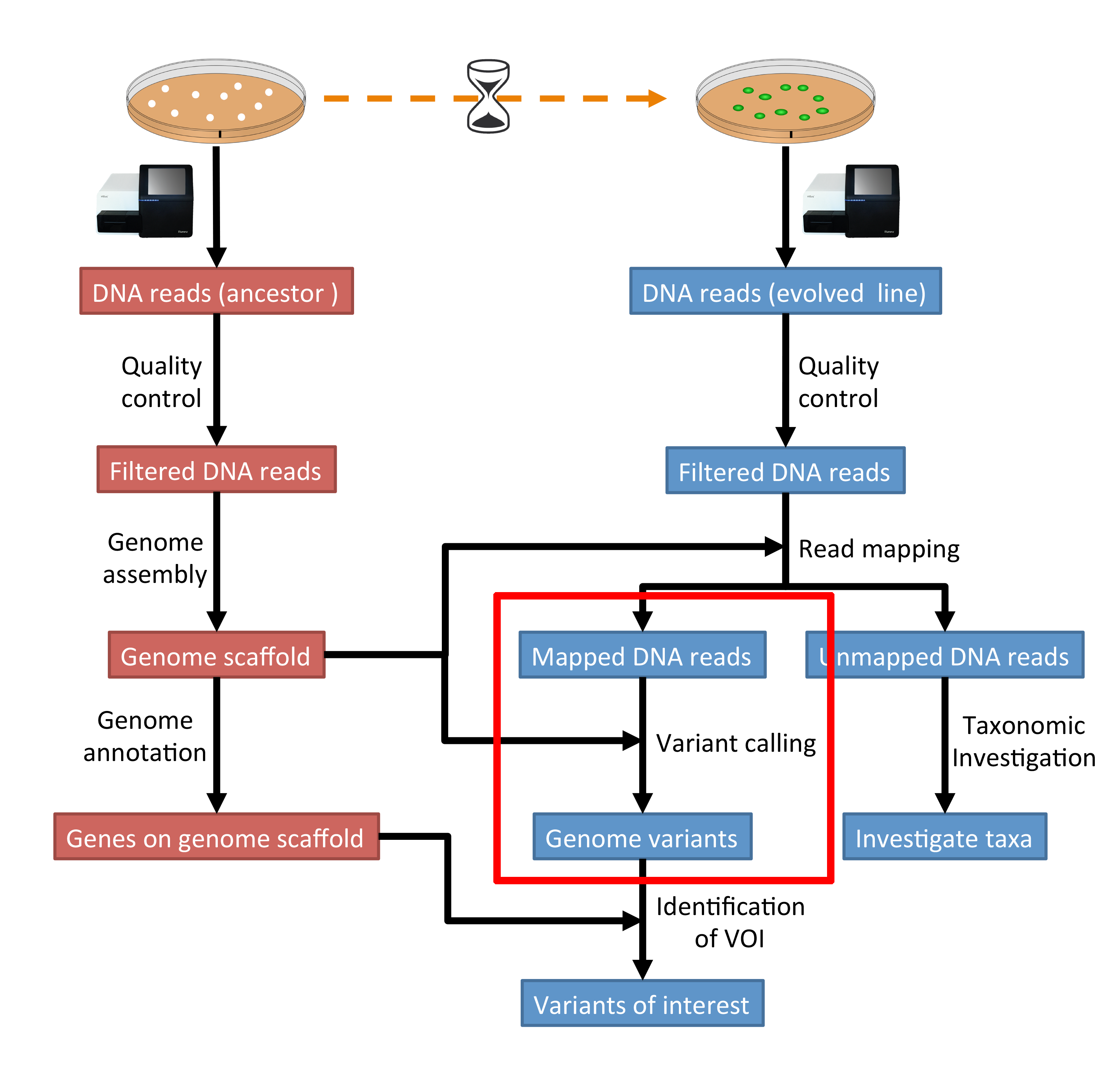
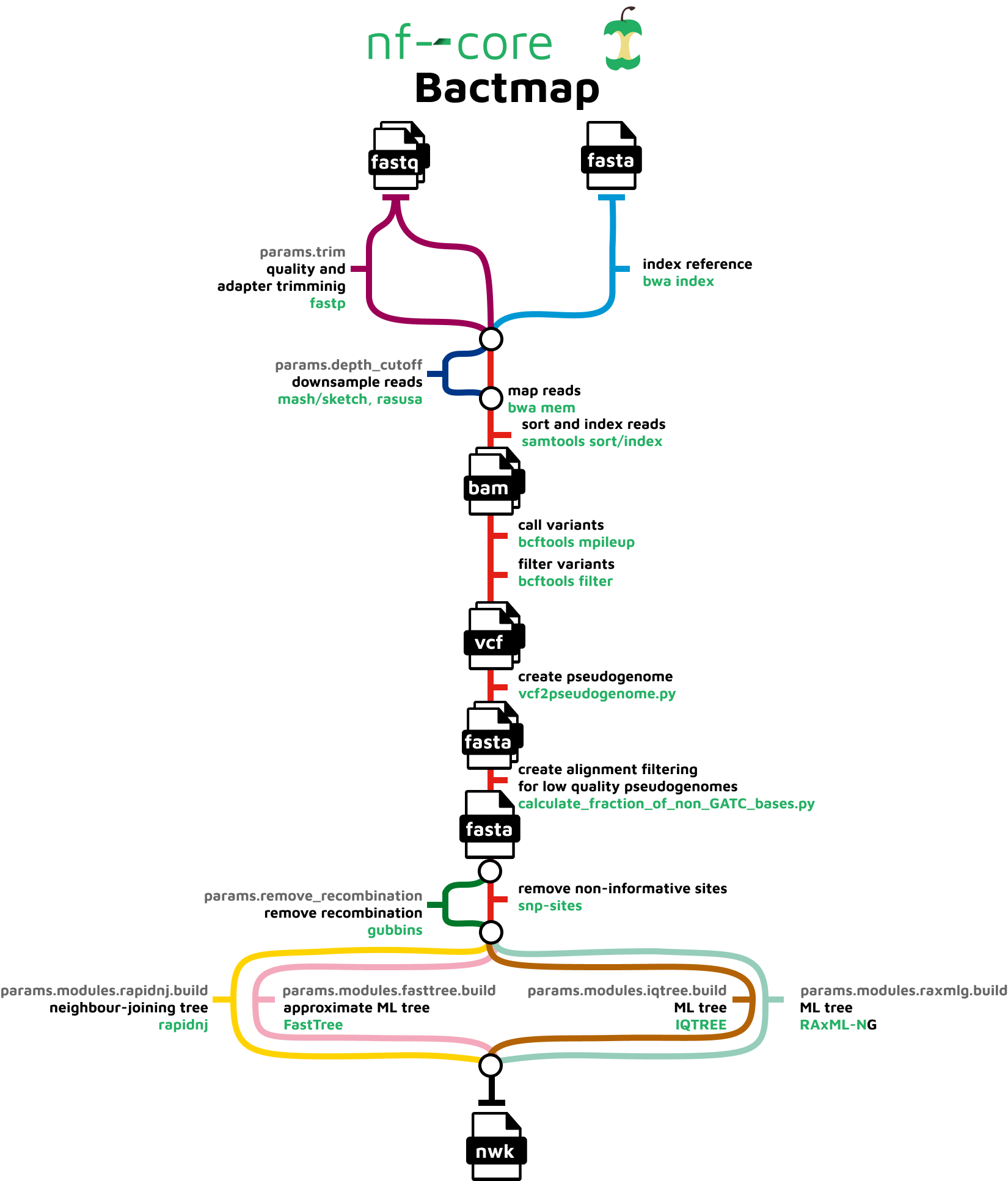
Variant calling using NGS and sequence capture data for population and evolutionary genomic inferences in Norway Spruce (Picea abies) | bioRxiv
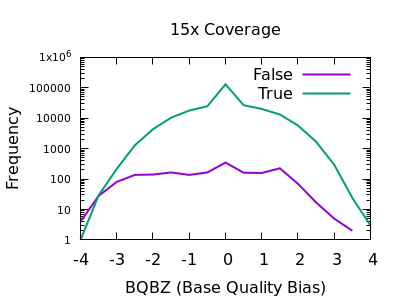
PLOS Genetics: No unexpected CRISPR-Cas9 off-target activity revealed by trio sequencing of gene-edited mice